Some people have noticed that some DNA frequencies work for some and don’t seem to work for others, and Spooky2 has pinpointed the reason for this, because of the way in which DNA, RNA and mRNA frequencies are calculated.
History background
Back in the 1930s, a brilliant scientist named Royal Rife designed electronic frequency devices that could disable pathogens and cure serious diseases like cancer and pneumonia. He found that each pathogen had a specific frequency of disability. To discover frequencies, Rife designed a very powerful microscope. Through careful observation, Rife can determine which frequencies disable the pathogen. He found that the frequency had to be precise and that other frequencies had no effect on the pathogen.
People often wonder why modern frequency therapy equipment is not as effective as these earliest machines, the reason is sad, Rife’s microscopes and electrotherapy equipment have not stood the test of time, most of them are damaged beyond repair, technical knowledge has been lost, we It is no longer possible to observe live pathogens at such high magnifications.
Without the right frequency, modern machines can’t match the amazing performance of the Royal Rife’s antique machines, Rife’s lifelong experiments proved that pathogens can be wiped out quickly by using the resonance frequency, Rife uses the term “absolutely coordinated resonance” to describe his frequency is how it works. 振”一詞來描述他的頻率是如何工作的。
Resonance – A playground swing is a very simple example of resonance, a light push at the right moment (frequency) will cause the child to swing higher and higher, the energy of each push adds kinetic energy to the swing, if the timing of the push Incorrect, resonance is lost and swing energy is reduced. Radio receivers also use resonance, which, when the radio is properly tuned, resonates with the frequency the radio station is transmitting.
Both DNA and RNA are genomes, and the genome holds the complete genetic information of an organism, providing all the information an organism needs to function. One of the main functions of DNA is to produce proteins, and resonance frequencies can prevent the genome from producing proteins and replicating, thereby disrupting the life function of cells. If a cell is a pathogen, it becomes nonviable and an easy target for our innate immune system.
DNA and RNA Antennas
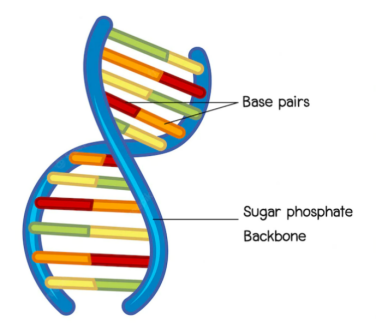
The genome forms biological single- and double-wire helical antennas tuned to specific frequencies, and applying those frequencies causes them to resonate. This idea is not new, a patent published about 15 years ago (Charlene A. Boehm, METHODS FOR DETERMINING THERAPEUTIC RESONANT FREQUENCIES, US 7,280,874 B2, October 9, 2007) describes how to derive from the axial length of DNA strands DNA resonance frequency. The resonant frequency of a normal-mode helical antenna depends on the radial length of the DNA, not the axial length. This is the length of the sugar-phosphate “backbone” shown in red below, and the resonance condition is satisfied if the radial length of the genomic sugar-phosphate “backbone” is the same as the applied wavelength. Antennas can be closed (circular genome) or open (linear genome).
Structural differences must be taken into account when calculating the resonance frequencies of RNA versus DNA genomes. RNA has an extra OH group on the sugar “backbone”, so the twists are tighter. The effective helix radius of RNA is also different from that of DNA, and these factors cause RNA to resonate at a different frequency than DNA, even though they have the same base pair count.
Genomic resonance frequencies can be calculated using the following mathematical formula:
Frequency = EMF propagation speed / effective genome length.
EMF propagation velocity is the speed at which electrical frequencies travel through a medium, the medium of which is the nucleoplasm for DNA and most RNAs. Genome resonance calculations must take into account the effective genome length. Genomes come in two basic configurations. Linear genomes are the most common. Calculations for this type of genome require subtracting one base pair from the total length, which is the effective base pair count (bn).
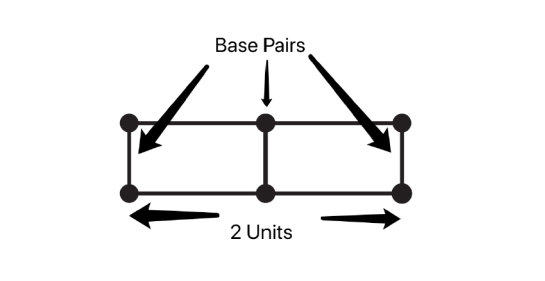
The figure above represents a linear double-stranded DNA genome with 3 base pairs with only 2 gaps between them, the total genome length is (number of bases – 1) x (distance between base pairs), a circular genome does not Base pairs need to be subtracted.
Genomic resonance occurs when the wavelength of the applied EMF signal is one octave of the genome length. An octave is a factor of 2, examples of octaves are 2, 4, 8, 16, and 32, EMF travels at the speed of light in a vacuum: 2.99792458E+8 meters per second, most of the genome is immersed in the nucleoplasm, where EMF propagation speed (vn) is slower: 1.23706749665013E+08 m/s.
Radial genome length can be calculated using the following formula:
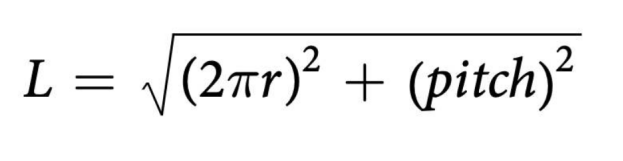
mRNA
The different sizes between DNA and RNA lead to different resonance frequencies, but there is another factor that must be considered, mRNA moves to the cytoplasm where proteins are made, and the EMF speed through the cytoplasm is 1.15164716490445E+08 m/s, so even efficient The base pair count is the same, and the resonance frequency of mRNA is different from that of DNA or “regular” RNA.
Working example
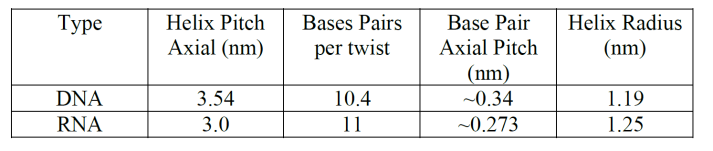
The table below shows a working example of DNA and RNA frequency derivation:
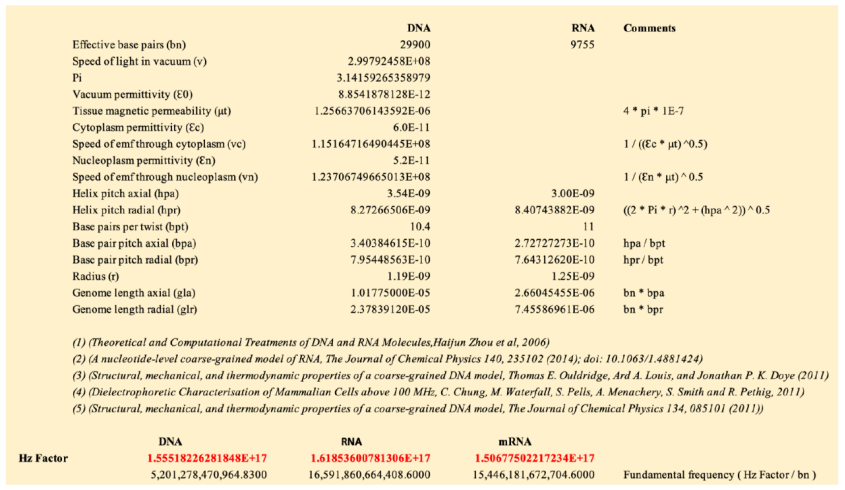
The resulting frequencies are very high, but can be reduced using selected subharmonics. The most common is to use sub-octave scales to align with standard antenna theory and practice, these are integer powers of 2, i.e. 2, 4, 8, 16, 32, etc.
Conclusion
Pathogens have 3 main types of genomes: DNA, RNA, and mRNA, which have different resonance frequencies even though they each have the same number of base pairs. By applying radio principles, the exact resonant frequency of the target genome can be calculated, and subharmonics of this frequency can then be applied to disrupt the pathogen’s vital functions.

